To illustrate Bayesian DCA on survival data, we load the example
dataset dca_survival_data shown below.
library(bayesDCA)
head(dca_survival_data)
#> outcomes.time outcomes.status model_predictions binary_test
#> 1 0.2948349 0.0000000 0.18806301 0
#> 2 1.8747960 0.0000000 0.25349061 1
#> 3 0.0355183 0.0000000 0.01025269 0
#> 4 1.2850960 0.0000000 0.04836840 0
#> 5 0.2398763 0.0000000 0.23502042 1
#> 6 2.0934150 0.0000000 0.04611183 0The dataset contains the time-to-event outcomes (a
survival::Surv object), the predictions from a prognostic
model, and the results from a binary prognostic test. The time horizon
for the event prediction is one time units (e.g., year), so we set
prediction_time = 1. We also set chains = 1 to
speed up MCMC sampling with Stan (in
practice you should use at least chains = 4, maybe with
cores = 4 for speed as well).
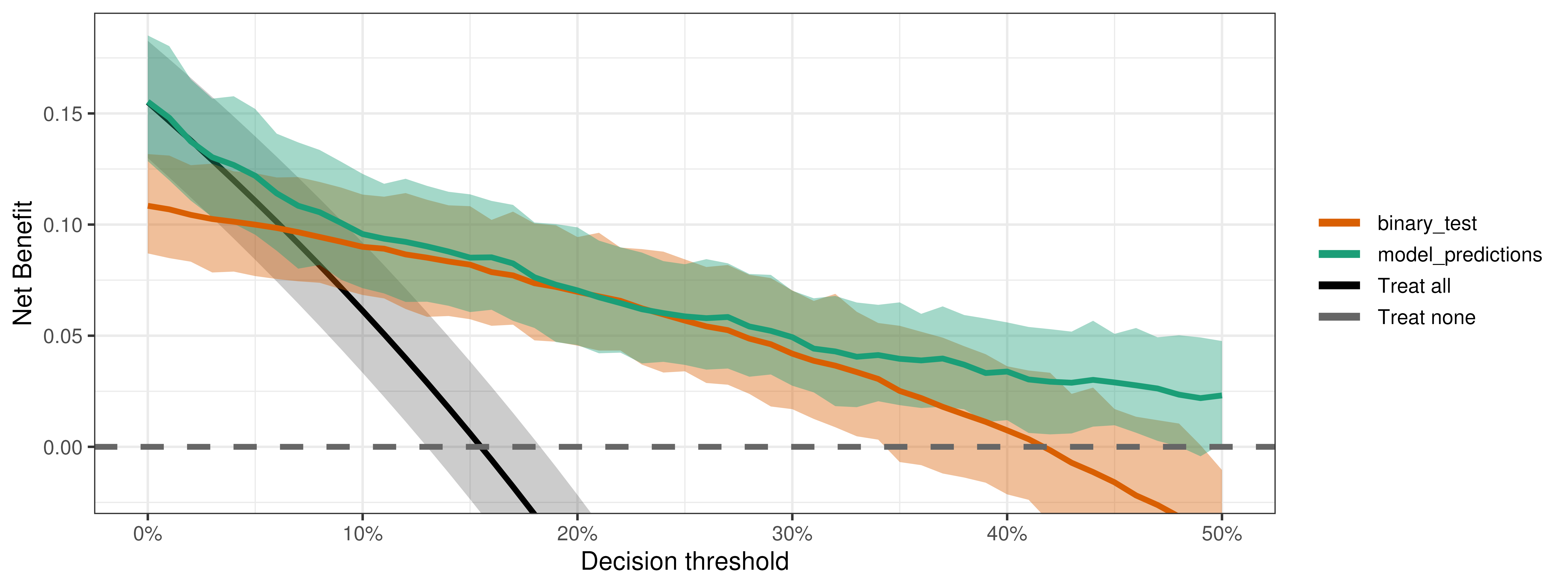
Interrogating the output
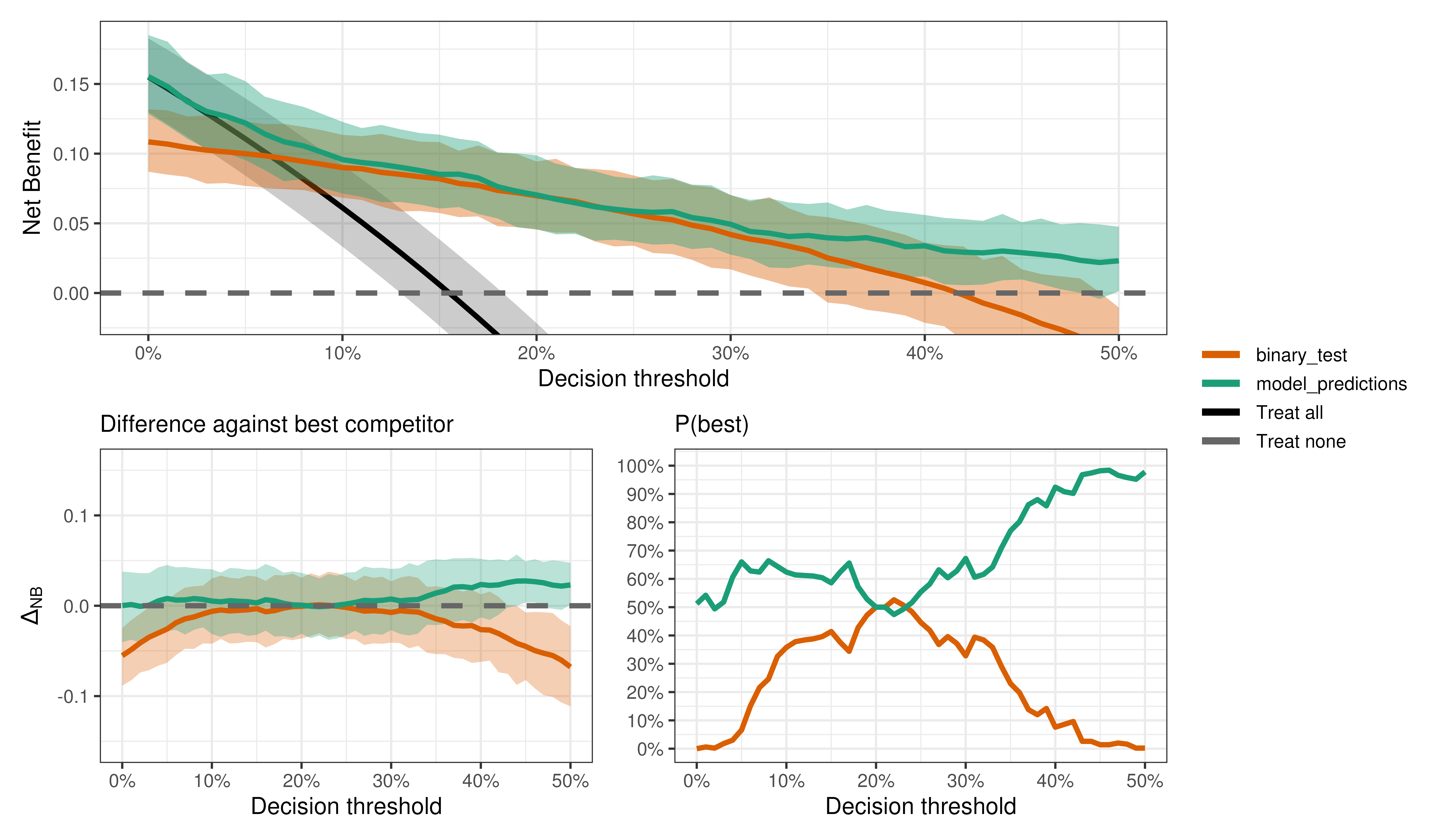
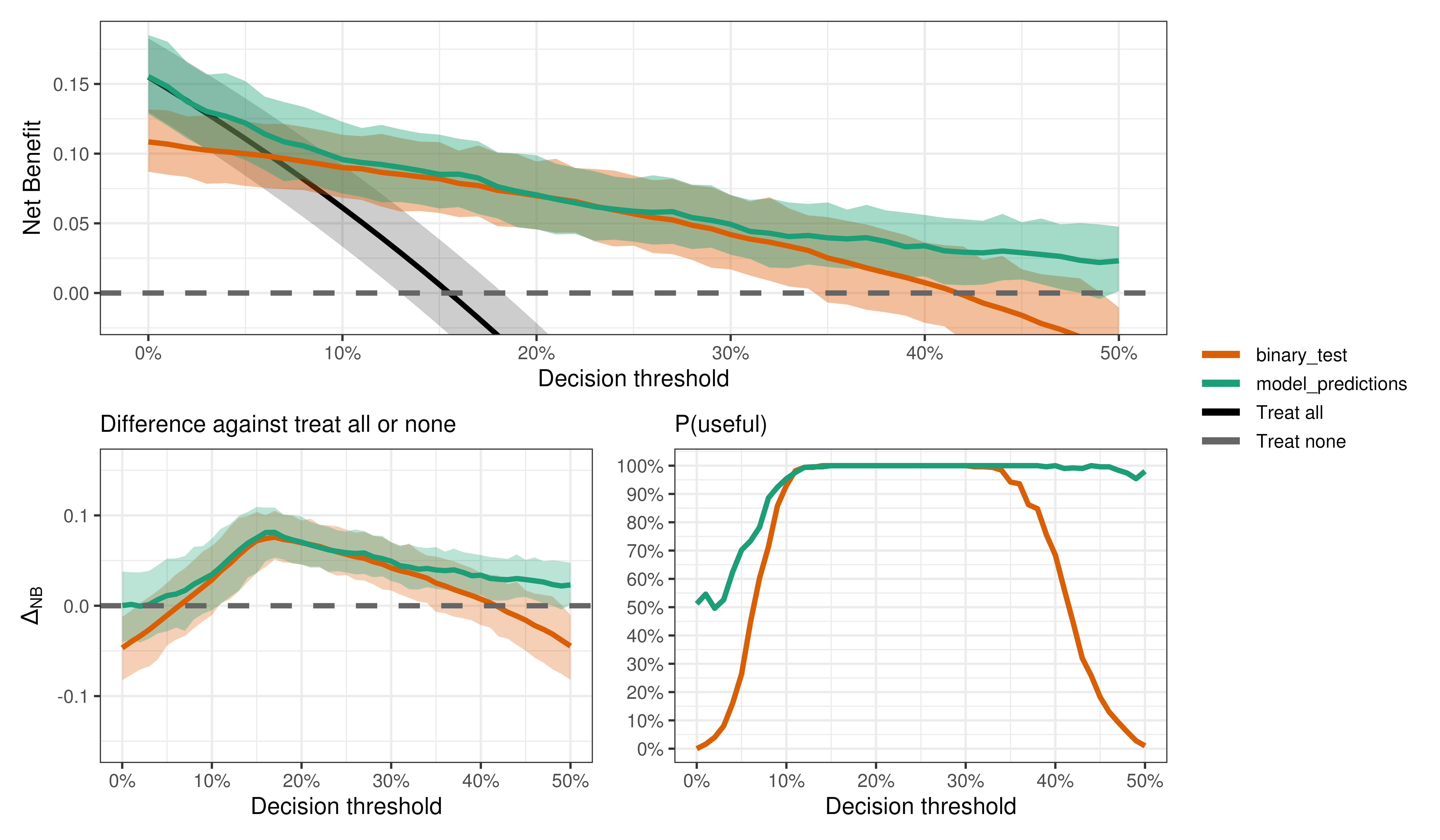
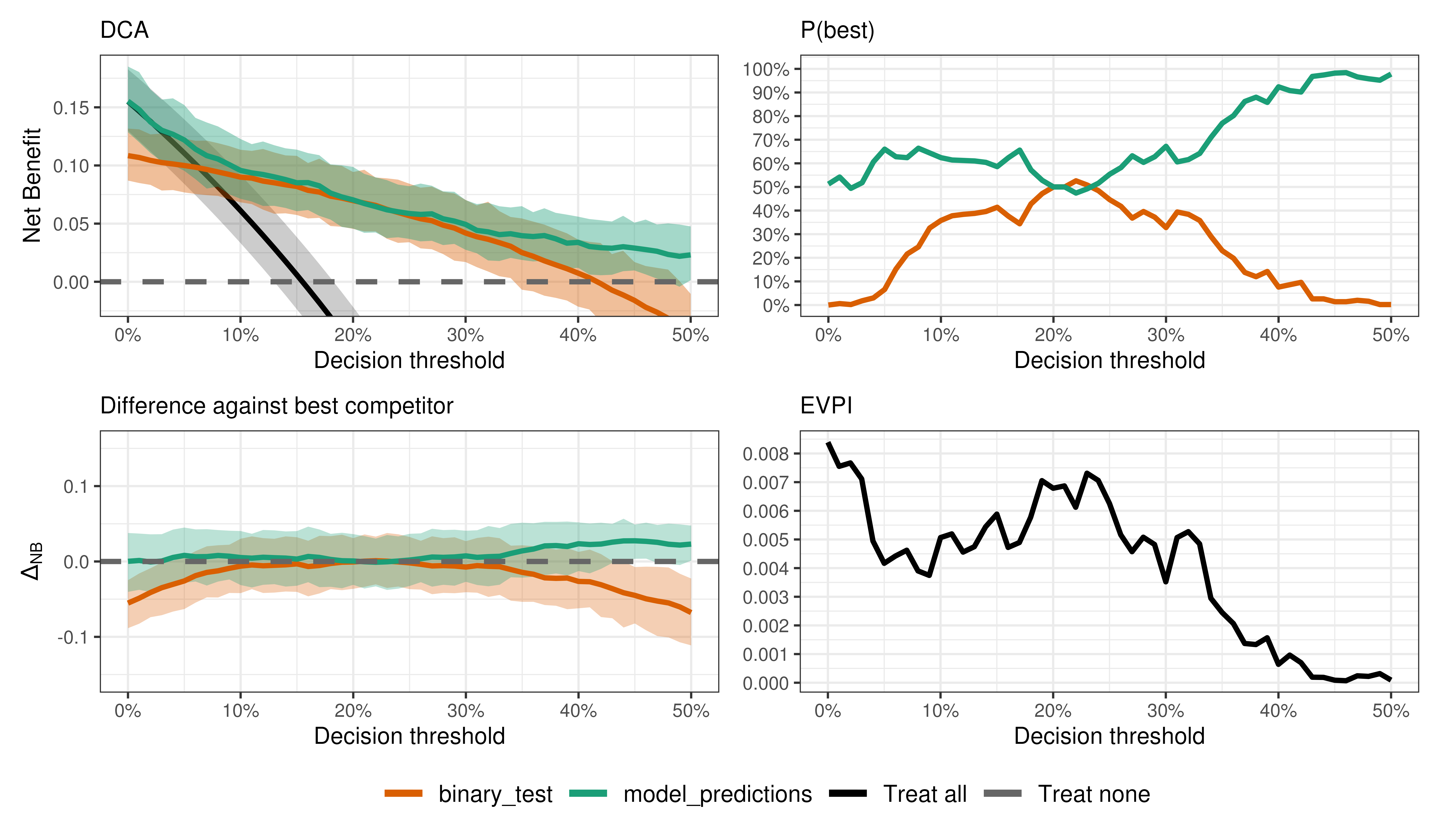
We can do all kinds of output interrogation just like with binary outcomes.
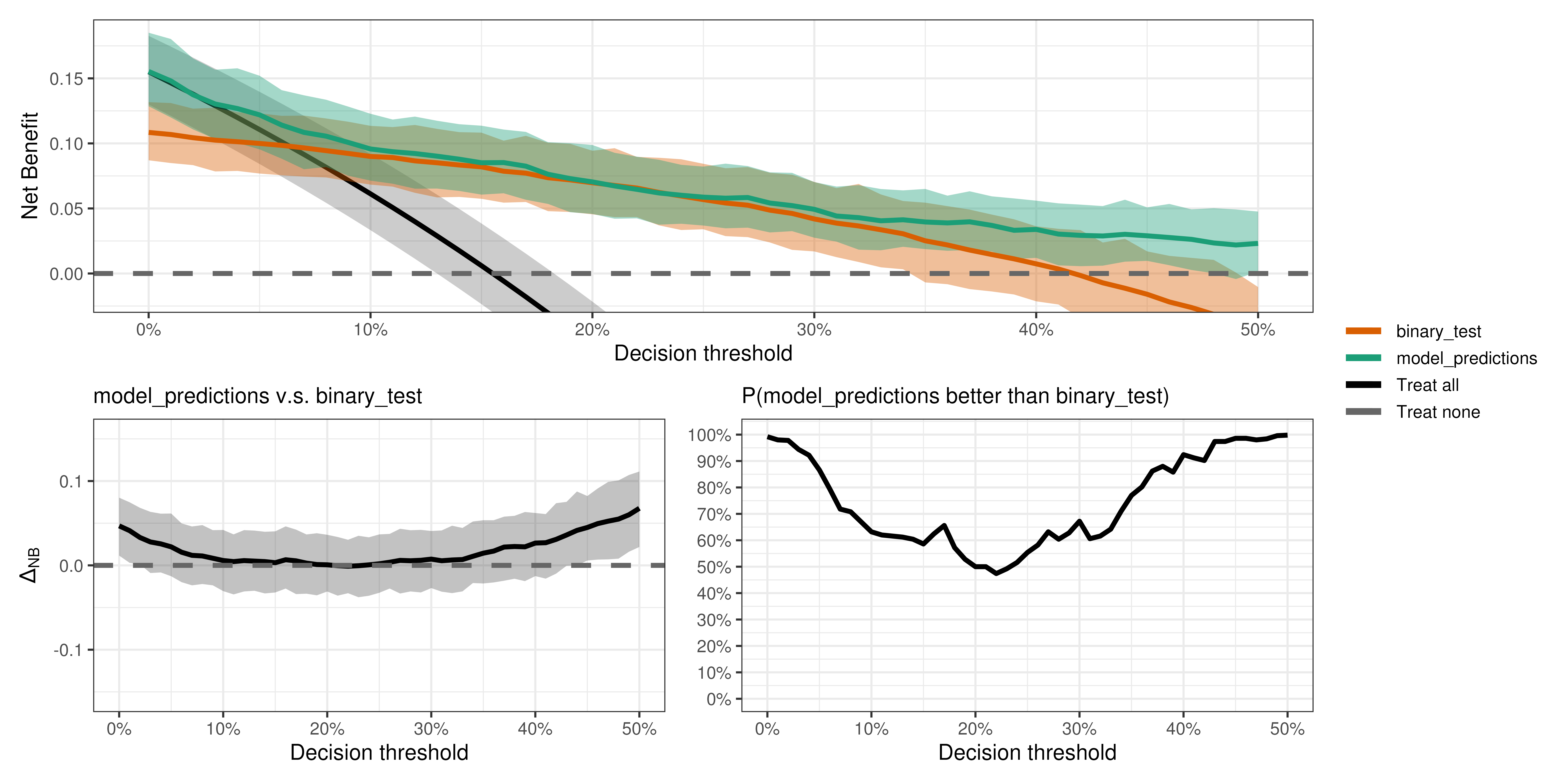
Is the model better than the test?
compare_dca(fit,
strategies = c("model_predictions", "binary_test"),
type = "pairwise")